R Programming Language: Dive into the world of data analysis and visualization with this comprehensive guide! We’ll explore R’s history, from its humble beginnings to its current status as a powerhouse in the data science field. Get ready to master its core features, manipulate data like a pro with dplyr, and create stunning visualizations using ggplot2. We’ll cover statistical modeling, working with external data, and even touch on the exciting world of machine learning.
Table of Contents
It’s gonna be a fun ride!
This guide provides a practical, hands-on approach to learning R, covering everything from basic syntax to advanced statistical modeling and machine learning techniques. We’ll walk through real-world examples, emphasizing best practices for data cleaning, analysis, and visualization. Whether you’re a complete beginner or have some prior programming experience, you’ll find this guide valuable in expanding your R skills and applying them to your own data projects.
History of R: R Programming Language
R’s journey from a niche statistical computing language to a dominant force in data science is a fascinating story of collaboration, innovation, and community growth. Its evolution reflects the changing landscape of data analysis and the ever-increasing demand for powerful and flexible tools.R’s origins lie in the 1990s, stemming from the S programming language developed at Bell Labs. This foundational influence shaped R’s core syntax and statistical capabilities, providing a robust base for future expansion.
The open-source nature of R, a key differentiator, fostered a collaborative environment that propelled its adoption and development.
R’s Development Timeline
The development of R can be understood through several key milestones. Initially conceived by Ross Ihaka and Robert Gentleman at the University of Auckland, New Zealand, the first version of R was released in 1995. This marked the beginning of a project that would transform the way data scientists and statisticians approach their work. Subsequent years saw steady improvements and the growth of a dedicated community contributing packages and extensions.
The formation of the R Core Team, a group of dedicated developers responsible for the core language, was crucial in maintaining consistency and guiding its development. Major releases, each bringing significant enhancements, have punctuated R’s history, reflecting the evolution of computing and data analysis techniques. The increasing adoption of R in academia and industry led to the creation of numerous user groups and conferences, further strengthening its community and influence.
Evolution of R’s Core Functionalities and Package Ecosystem
Initially, R’s core functionality focused on statistical computing tasks, providing a rich set of tools for data manipulation, statistical modeling, and visualization. However, over time, its capabilities have broadened significantly. The development of the CRAN (Comprehensive R Archive Network), a central repository for R packages, has been instrumental in this expansion. The package ecosystem, built by a global community of developers, offers solutions for a vast array of tasks, extending R’s reach into machine learning, data mining, web development, and many other areas.
This community-driven development is a defining characteristic of R, distinguishing it from proprietary alternatives. The sheer number and diversity of packages available highlight the flexibility and adaptability of the language.
Performance Improvements in R
Early versions of R were known for their relatively slower performance compared to some compiled languages. This was partly due to its interpreted nature and the limitations of computing resources at the time. However, significant performance improvements have been made over the years. Optimizations to the core language, the development of just-in-time (JIT) compilers such as those incorporated in projects like Rcpp, and the increasing availability of powerful computing hardware have all contributed to a substantial increase in R’s speed and efficiency.
Modern R is capable of handling datasets of significant size and complexity with reasonable performance, thanks to these advancements and the sophisticated algorithms implemented within various packages. For instance, the performance gains in handling large datasets using data.table package are a significant improvement over base R functionality. This enhanced performance has made R a viable option for a wider range of applications, including those demanding real-time or high-throughput processing.
Core Features and Syntax
R’s power lies not just in its statistical capabilities but also in its elegant and expressive syntax, coupled with robust object-oriented features. This allows for efficient data manipulation, analysis, and the creation of reproducible research. Understanding these core elements is key to unlocking R’s full potential.R’s object-oriented features are based on S3 and S4 systems, offering flexibility in how you structure and work with data.
While S3 is simpler and more commonly used, S4 provides a more formal and rigorous approach. These systems allow for methods to be dispatched based on the class of an object, promoting code reusability and maintainability. For example, a generic `plot()` function can be used to visualize different data types (vectors, matrices, data frames) by calling the appropriate method specific to each class.
Object-Oriented Programming in R
R’s object-oriented programming (OOP) features, primarily S3 and S4, enable creating reusable and extensible code. S3 is simpler, using a convention of adding class attributes to objects and defining generic functions that dispatch methods based on the class. S4 is more formal, explicitly defining classes and methods. A practical application is creating custom data structures and associated functions for specific analysis needs.
Imagine building a class for “gene expression data” with methods for normalization, filtering, and visualization. This would avoid repetitive coding and improve code organization. This approach is invaluable in larger projects where consistency and maintainability are critical.
Base Functions for Data Manipulation and Analysis
R boasts a rich set of base functions that provide the building blocks for data manipulation and analysis. Functions like `subset()`, `aggregate()`, `apply()`, `lapply()`, `sapply()`, `tapply()`, and `merge()` are fundamental for tasks ranging from simple data filtering and aggregation to more complex data transformations and joining. For instance, `subset()` allows for efficient filtering of data frames based on specified criteria.
So, I’m totally into R programming right now, especially for data visualization. I’ve been generating a ton of PNG files with my plots, and honestly, it’s a pain to send them all individually. That’s why I’ve been using a super helpful png to pdf converter to easily compile them into a single PDF for sharing. It saves me a ton of time, letting me get back to the fun part – more R coding!
`apply()` and its family of functions provide powerful ways to apply functions across rows, columns, or other dimensions of data structures. These functions are frequently used in conjunction with other base functions and packages, making them essential for almost any R workflow. For example, you might use `subset()` to select specific rows, then `aggregate()` to calculate summary statistics for subgroups.
R Data Structures
Understanding R’s data structures is crucial for effective programming. Each structure has unique characteristics and is best suited for different types of data and analyses.
| Data Structure | Description | Example | Use Case |
|---|---|---|---|
| Vector | Ordered sequence of elements of the same type. | my_vector <- c(1, 2, 3, 4, 5) |
Storing a series of measurements or observations. |
| Matrix | Two-dimensional array of elements of the same type. | my_matrix <- matrix(1:9, nrow = 3) |
Representing tabular data or images. |
| List | Ordered collection of elements of potentially different types. | my_list <- list(name = "Alice", age = 30, scores = c(85, 92, 78)) |
Storing heterogeneous data, such as a combination of text, numbers, and vectors. |
| Data Frame | Table-like structure where each column can have a different data type. | my_data_frame <- data.frame(name = c("Alice", "Bob"), age = c(30, 25), scores = c(85, 90)) |
Storing and analyzing tabular data, often used in statistical analysis. |
Data Manipulation with dplyr

dplyr is a game-changer for data manipulation in R. It provides a consistent and intuitive grammar for data wrangling, making complex operations much easier to read and write than using base R functions. This section will explore some key dplyr verbs and demonstrate how to build robust data manipulation workflows.
dplyr's core strength lies in its set of verbs, each designed for a specific data manipulation task. These verbs operate on data frames, transforming them in predictable ways. The beauty of dplyr is its elegance and efficiency, particularly when dealing with large datasets where base R's approaches can become cumbersome and slow.
dplyr Verbs: Examples of Data Cleaning and Transformation
The six most commonly used dplyr verbs are select(), filter(), mutate(), summarize(), arrange(), and group_by(). Let's illustrate their use with a simple example. Imagine we have a data frame called `my_data` containing information about students: their ID, name, grade, and age.
Suppose we want to select only the ID and name columns. We would use select():
selected_data <- select(my_data, ID, name)
To filter students older than 15, we use filter():
older_students <- filter(my_data, age > 15)
To add a new column calculating the student's age in years from their birthdate (assuming we have a birthdate column), we use mutate():
my_data <- mutate(my_data, age_years = as.numeric(difftime(Sys.Date(), birthdate, units = "days"))/365.25)
To summarize the average grade of all students, we use summarize():
average_grade <- summarize(my_data, mean_grade = mean(grade, na.rm = TRUE))
To sort the data by grade, from highest to lowest, we use arrange():
sorted_data <- arrange(my_data, desc(grade))
Finally, to group data by grade and then calculate the average age within each grade group, we use group_by() along with summarize():
grade_age_summary <- my_data %>% group_by(grade) %>% summarize(mean_age = mean(age, na.rm = TRUE))
Workflow for Data Manipulation using dplyr with Error Handling
A robust dplyr workflow should incorporate error handling to prevent unexpected crashes. This can involve checking for missing data, handling incorrect data types, and gracefully managing errors that might occur during data transformations.
A typical workflow might involve the following steps:
- Data Import and Inspection: Begin by importing your data and inspecting it for missing values, incorrect data types, or inconsistencies. Use functions like
summary(),str(), andis.na()to check your data's structure and identify potential problems. - Data Cleaning: Address any identified issues. This might involve imputation of missing values (using functions like
imputeTS::na_interpolation()), data type conversion (using functions likeas.numeric()oras.factor()), or removal of outliers. - Data Transformation: Apply dplyr verbs to reshape and transform the data according to your analysis needs. This is where you use
select(),filter(),mutate(), etc. - Error Handling within Transformations: Use
tryCatch()to handle potential errors during transformations. For example, if a calculation might result in division by zero, wrap it in atryCatch()block to handle the error gracefully. - Data Validation: After transformations, validate the data to ensure the results are as expected. Check data ranges, distributions, and summaries to detect any anomalies.
Benefits of dplyr over Base R for Data Manipulation
dplyr offers several advantages over base R for data manipulation:
Firstly, dplyr's syntax is much more readable and intuitive. The use of verbs like select(), filter(), etc., makes the code easier to understand and maintain. Base R's approach often involves a sequence of nested functions which can be difficult to follow.
Secondly, dplyr is generally more efficient, particularly for larger datasets. Its optimized functions often outperform base R's equivalents in terms of speed. This is especially important when working with massive datasets where performance is crucial.
Finally, dplyr promotes a more consistent and organized approach to data manipulation. The consistent grammar makes it easier to write reproducible and maintainable code, reducing the likelihood of errors and making collaboration easier.
Data Visualization with ggplot2
ggplot2 is the gold standard for data visualization in R. It's based on the "grammar of graphics," a powerful conceptual framework that lets you build complex visualizations by layering different components. This approach makes it incredibly flexible and allows for highly customized plots, far beyond what you'd find in simpler plotting functions. We'll explore its capabilities and compare it to other visualization libraries.
Building a Complex Visualization with ggplot2: A Step-by-Step Guide
Let's create a complex visualization showing the relationship between carat weight, price, and cut quality of diamonds using the `diamonds` dataset included in ggplot2. We'll build this plot step-by-step, demonstrating the power of ggplot2's layered approach.First, we load the necessary library: `library(ggplot2)`. Then, we start with the base layer: `ggplot(data = diamonds, aes(x = carat, y = price, color = cut))`.
This sets the data, x and y axes, and color aesthetic. Next, we add points: `+ geom_point()`. This creates a scatter plot showing the relationship between carat and price, with points colored by cut quality. To enhance readability, we can add a smoother: `+ geom_smooth(method = "lm")`. This overlays a linear regression line, showing the overall trend.
Finally, we can improve the aesthetics with a title and axis labels: `+ labs(title = "Diamond Price vs. Carat Weight", x = "Carat", y = "Price", color = "Cut Quality") + theme_bw()`. The `theme_bw()` function provides a clean black and white theme. The complete code would be: library(ggplot2)ggplot(data = diamonds, aes(x = carat, y = price, color = cut)) + geom_point() + geom_smooth(method = "lm") + labs(title = "Diamond Price vs. Carat Weight", x = "Carat", y = "Price", color = "Cut Quality") + theme_bw()This produces a sophisticated visualization displaying the relationship between carat, price, and cut quality, clearly illustrating the trends and patterns in the data.
Comparison of ggplot2 with Other Visualization Libraries
ggplot2's grammar of graphics distinguishes it from other R visualization libraries like base R graphics and lattice. Base R graphics are procedural, requiring many individual function calls to create a plot. Lattice provides a structured approach but lacks the flexibility and customization options of ggplot2. ggplot2's declarative nature, where you specify what you want the plot to look like rather than how to create it step-by-step, simplifies complex visualizations and makes them easier to maintain and modify.
Python's matplotlib and seaborn offer similar functionalities, with seaborn's high-level functions mirroring some of ggplot2's ease of use, but ggplot2 remains a powerful and popular choice within the R ecosystem.
Three Visualizations of the Same Dataset
Let's use the `diamonds` dataset again to illustrate three different visualizations using ggplot2, highlighting their strengths and weaknesses.First, we'll use a boxplot to compare the price distribution across different cut qualities: ggplot(diamonds, aes(x = cut, y = price)) + geom_boxplot() + labs(title = "Diamond Price Distribution by Cut Quality")This visualization effectively shows the median, quartiles, and outliers for price within each cut category. However, it doesn't show the relationship between price and carat weight.Next, a histogram will show the distribution of diamond carat weights: ggplot(diamonds, aes(x = carat)) + geom_histogram(bins = 30, fill = "lightblue", color = "black") + labs(title = "Distribution of Diamond Carat Weights")This effectively displays the frequency distribution of carat weights.
The weakness here is that it doesn't show the relationship with price or cut quality.Finally, a scatter plot with facets will show the relationship between price and carat weight, broken down by cut quality: ggplot(diamonds, aes(x = carat, y = price)) + geom_point(alpha = 0.5) + facet_wrap(~ cut) + labs(title = "Diamond Price vs. Carat Weight by Cut Quality")This visualization combines the strengths of both the scatter plot and the facetting approach. It shows the relationship between price and carat, but also allows for comparison across different cut qualities.
The `alpha = 0.5` adds transparency to handle overplotting. The choice of visualization depends on the specific question being asked and the aspects of the data you want to emphasize.
Statistical Modeling in R

R's robust statistical capabilities make it a go-to tool for data analysis and modeling. From simple linear regressions to complex mixed-effects models, R offers a comprehensive suite of functions and packages to fit, interpret, and diagnose various statistical models. This section will explore some key aspects of statistical modeling in R, focusing on linear regression and hypothesis testing.
Linear Regression in R
Linear regression aims to model the relationship between a dependent variable and one or more independent variables. In R, the `lm()` function is the workhorse for fitting linear regression models. Let's consider a simple example where we want to predict house prices (dependent variable) based on their size (independent variable). Assuming we have a dataset called `house_data` with columns `price` and `size`, we can fit a linear model like this: model <- lm(price ~ size, data = house_data)
summary(model)
The `summary()` function provides key information, including the estimated coefficients (intercept and slope), their standard errors, t-values, p-values, and R-squared.
The R-squared value indicates the proportion of variance in the dependent variable explained by the model. The p-values associated with the coefficients test the null hypothesis that the coefficient is equal to zero. A low p-value (typically less than 0.05) suggests a statistically significant relationship between the independent and dependent variables.To visualize the model's fit, we can create a scatter plot of the data with the regression line overlaid: plot(house_data$size, house_data$price, main = "House Price vs. Size", xlab = "Size", ylab = "Price")abline(model, col = "red")This plot shows the observed data points and the best-fitting line according to our model.
Diagnostic plots, generated using `plot(model)`, can help assess the model's assumptions (linearity, constant variance, normality of residuals). Deviations from these assumptions might indicate the need for model adjustments or alternative approaches. For instance, if the residual plot shows a non-constant variance (heteroscedasticity), weighted least squares regression might be more appropriate. If the residuals are not normally distributed, transformations of the variables or a different model might be considered.
Hypothesis Testing in R
Hypothesis testing involves using sample data to make inferences about a population. R provides numerous functions for conducting various hypothesis tests. Let's say we want to test the hypothesis that the average price of houses is different from a specific value (e.g., $500,000). We can use a one-sample t-test: t.test(house_data$price, mu = 500000)The output provides the t-statistic, degrees of freedom, and p-value.
If the p-value is below the significance level (e.g., 0.05), we reject the null hypothesis and conclude that the average house price is significantly different from $500,000. Other common hypothesis tests readily available in R include ANOVA (`aov()`), chi-squared tests (`chisq.test()`), and non-parametric tests like the Wilcoxon test (`wilcox.test()`). The choice of test depends on the data type and research question.
Comparison of Statistical Models
Linear regression is suitable for modeling the relationship between a continuous dependent variable and one or more independent variables (continuous or categorical). Logistic regression, on the other hand, is used when the dependent variable is binary (0 or 1). For example, predicting whether a customer will click on an ad (binary outcome) based on their demographics and browsing history would require logistic regression.
The `glm()` function in R, with the `family = binomial` argument, is used for fitting logistic regression models.
The choice of statistical model depends critically on the nature of the data and the research question. Understanding the assumptions and limitations of each model is crucial for accurate and meaningful analysis.
Other models, such as Poisson regression (for count data), survival analysis models (for time-to-event data), and more advanced techniques like generalized additive models (GAMs) and mixed-effects models, offer further flexibility in handling different types of data and research questions. R provides extensive support for all these models through various packages.
Working with External Data
Okay, so you've got your R skills up to speed, but where's the data? Real-world analysis means getting your hands dirty with external datasets. This section covers importing data from various sources, cleaning it up, and prepping it for analysis – all crucial steps before you can start crunching numbers. Think of this as the pre-game warm-up before the main event.Getting data into R is the first hurdle.
Luckily, R offers a ton of tools to handle various file formats and databases. We'll focus on some of the most common ones: CSV files, Excel spreadsheets, and SQL databases. Proper data import is key to avoiding headaches later on.
Importing Data from CSV Files
CSV (Comma Separated Values) files are a super common way to share data. They're simple text files, easily readable by humans and machines. In R, the `read.csv()` function is your best friend. For example, `my_data <- read.csv("my_file.csv")` reads the data from "my_file.csv" and stores it in a data frame called `my_data`. You can specify options like the separator (if it's not a comma) or the header row using additional arguments within the function. For instance, if your file uses semicolons as separators, you would use `read.csv("my_file.csv", sep = ";")`. Always check your data after importing to ensure it's been read correctly.
Importing Data from Excel Files
Excel files (.xls or .xlsx) are ubiquitous, especially in business and research. The `readxl` package provides the `read_excel()` function, which makes importing data from Excel a breeze.
First, you'll need to install the package using `install.packages("readxl")` and then load it with `library(readxl)`. Then, `my_data <- read_excel("my_excel_file.xlsx", sheet = "Sheet1")` reads data from the first sheet ("Sheet1") of "my_excel_file.xlsx". Remember to specify the sheet name if you need data from a different sheet.
Importing Data from SQL Databases
SQL databases are powerful for managing large datasets. R can connect to various SQL databases (like MySQL, PostgreSQL, or SQLite) using packages like `RMySQL`, `RPostgres`, or `RSQLite`. These packages allow you to execute SQL queries directly from R and import the results into a data frame.
For example, after installing and loading the necessary package (e.g., `library(RMySQL)`), you might use a function like `dbGetQuery()` to execute a query and retrieve the data. The specific syntax depends on the database system.
Handling Missing Data
Missing data is a fact of life in real-world datasets. R provides several ways to handle this. The simplest approach is to remove rows or columns with missing values using functions like `na.omit()`. However, this can lead to a loss of information. A more sophisticated approach is to impute missing values using methods like mean imputation (replacing missing values with the mean of the column), median imputation, or more advanced techniques available in packages like `mice` (Multivariate Imputation by Chained Equations).
The choice of method depends on the nature of your data and the type of analysis you're performing. Carefully consider the implications of your chosen method.
Handling Outliers
Outliers are data points that significantly deviate from the rest of the data. They can skew your analysis and lead to misleading results. Identifying outliers often involves visual inspection (using box plots or scatter plots) or statistical methods like calculating z-scores. Once identified, you can choose to remove them, transform the data (e.g., using logarithmic transformation), or use robust statistical methods that are less sensitive to outliers.
There is no single "best" method; the optimal approach depends on the context and your goals.
Data Cleaning and Preprocessing Workflow
A typical data cleaning and preprocessing workflow might look something like this:
- Import Data: Use the appropriate function based on your data source (CSV, Excel, SQL, etc.).
- Inspect Data: Use functions like `head()`, `summary()`, `str()`, and visualizations to understand your data's structure, identify missing values, and spot potential outliers.
- Handle Missing Data: Decide on an imputation method or decide whether to remove rows/columns with missing values.
- Handle Outliers: Identify and address outliers using appropriate methods.
- Data Transformation: Consider transformations (e.g., scaling, standardization, log transformation) to improve the performance of your statistical models.
- Data Cleaning: Remove unnecessary columns, correct inconsistencies, and ensure data types are correct.
This workflow is iterative; you may need to revisit earlier steps as you discover new issues. Remember that thorough data cleaning is essential for reliable analysis.
R Packages and Extensions
R's power lies not just in its core functionality, but in its vast ecosystem of packages. These packages, essentially collections of functions and data, extend R's capabilities dramatically, allowing users to tackle almost any data science task imaginable. Think of them as add-ons that supercharge your R experience, providing specialized tools for everything from machine learning to web scraping.R's package system is incredibly efficient and well-maintained, making it easy to find, install, and use new tools.
This extensibility is a key reason for R's enduring popularity in the data science community. Let's explore some key packages and how they compare.
Essential R Packages for Data Science
Five essential packages for data science in R are `dplyr`, `ggplot2`, `tidyr`, `caret`, and `stringr`. These packages cover a wide range of tasks, from data manipulation and visualization to machine learning and text analysis. Their combination provides a powerful toolkit for a data scientist.
- `dplyr`: Provides a grammar of data manipulation, offering intuitive functions for filtering, selecting, summarizing, and mutating data frames. It simplifies data wrangling tasks significantly.
- `ggplot2`: A powerful and flexible system for creating elegant and informative data visualizations. It uses a layered grammar of graphics, allowing users to build complex plots from simpler components.
- `tidyr`: Complements `dplyr` by providing functions for tidying data. This means transforming data into a consistent and usable format, often involving reshaping and restructuring data frames.
- `caret`: A comprehensive package for classification and regression training. It provides a unified interface to many machine learning algorithms, simplifying model training, tuning, and evaluation.
- `stringr`: Offers a consistent set of functions for working with strings. This is crucial for tasks like text cleaning, manipulation, and analysis, which are common in data science projects.
Comparative Analysis of Packages for Data Manipulation
Several packages offer similar functionalities for data manipulation, but with different approaches. Let's compare `dplyr`, `data.table`, and base R's built-in functions.
While base R offers functions like `subset()`, `aggregate()`, and `apply()`, they can be less intuitive and require more lines of code than `dplyr` for complex operations. `data.table` is known for its speed and efficiency, especially when dealing with very large datasets. However, its syntax can be steeper learning curve compared to the more user-friendly `dplyr`. The choice often depends on the dataset size, the complexity of the manipulation, and the user's familiarity with each package.
| Package | Strengths | Weaknesses |
|---|---|---|
| `dplyr` | Intuitive syntax, excellent for data wrangling, good for interactive exploration | Can be slower than `data.table` for very large datasets |
| `data.table` | Extremely fast, efficient for large datasets | Steeper learning curve, syntax can be less readable |
| Base R | Familiar to R users, readily available | Can be less concise and more complex for advanced operations |
Installing and Loading R Packages
Installing and loading R packages is a straightforward process. To install a package, you use the `install.packages()` function, specifying the package name as a character string. For example, to install the `dplyr` package, you would type:
install.packages("dplyr")
Once installed, you need to load the package into your current R session using the `library()` function. This makes the package's functions available for use. For example:
library(dplyr)
If a package isn't installed, R will throw an error message. Always remember to install a package before attempting to load it. Many R packages are hosted on CRAN (the Comprehensive R Archive Network), a central repository of R software. Others may be found on GitHub or other repositories. The `install.packages()` function handles downloading and installing from various sources.
Reproducible Research with R Markdown
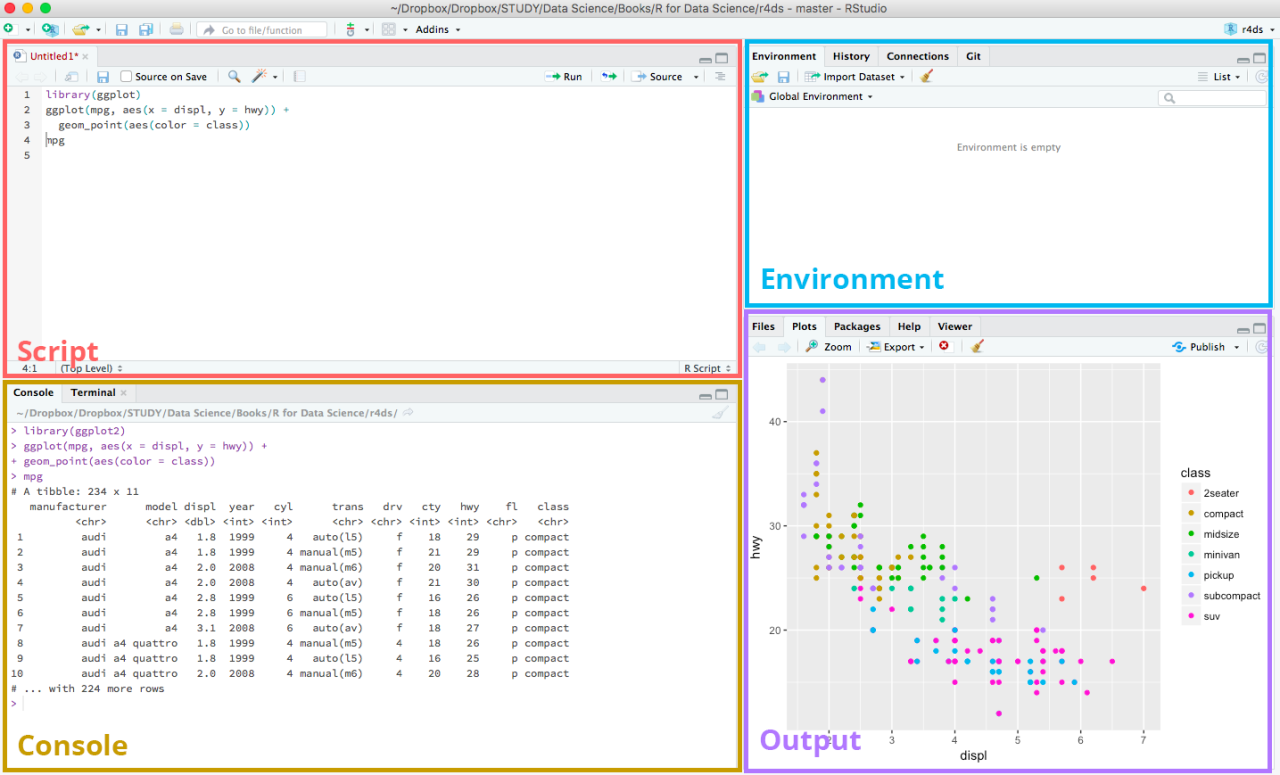
R Markdown is your secret weapon for creating reproducible research reports and presentations. It seamlessly blends your R code, its output (including stunning visualizations!), and your insightful narrative into a single, cohesive document. This allows others (and your future self!) to easily understand your analysis, reproduce your results, and even build upon your work. Think of it as a supercharged lab notebook that's also publication-ready.
The beauty of R Markdown lies in its simplicity and power. You write your analysis using a straightforward markdown syntax, embedding R code chunks within the text. When you knit the document, R Markdown executes the code, generates the output, and integrates everything into a polished final product – be it a PDF report, a Word document, a presentation, or even a website.
This ensures that your analysis is not only well-documented but also completely reproducible.
Designing an R Markdown Document
An R Markdown document typically consists of several key elements. First, you have the YAML header, which provides metadata like the document's title, author, and output format. Next comes the body of the document, where you'll use markdown syntax for text formatting and embed R code chunks using the ````r` and ````` delimiters. Within these code chunks, you can write your R code, generate plots, and create tables.
Finally, the knitted document combines all these elements into a single, cohesive output. For example, a simple R Markdown document might start with a YAML header specifying the title, author, and output format (e.g., PDF), followed by a markdown section introducing the analysis, and then an R code chunk to load and analyze a dataset. Subsequent sections would incorporate additional R code chunks and narrative text, building the complete analysis.
Using R Markdown for Reports and Presentations
R Markdown excels at creating both comprehensive research reports and engaging presentations. For reports, you can leverage its ability to integrate code, output, and narrative text to provide a detailed and transparent account of your analysis. The output can be exported to various formats, including PDF, Word, and HTML, making it easy to share your findings with a wide audience.
For presentations, R Markdown offers the `revealjs` output format, which generates interactive slideshows. You can include code chunks that dynamically update visualizations as you progress through the presentation, making it a powerful tool for data storytelling. Imagine presenting a business report, where you dynamically update charts based on user inputs during your presentation! This is easily achievable with R Markdown and `revealjs`.
Best Practices for Reproducible Research
Reproducibility is paramount in scientific research. R Markdown significantly enhances this by ensuring that your analysis is completely transparent and easily replicable. Here are some best practices:
First, always clearly define your research question and objectives. Then, meticulously document your data sources and any data preprocessing steps. Use descriptive variable names in your code, and add comments to explain complex logic. For any external data used, specify the exact source and version. Include a clear and concise explanation of your analysis methods.
This ensures transparency and allows others to understand and reproduce your findings.
Furthermore, always version control your R Markdown document and any associated data files using a system like Git. This allows you to track changes over time and easily revert to previous versions if needed. Consider creating a project directory that contains all your code, data, and documentation. This makes it easier to manage your project and share it with collaborators.
Finally, use well-structured code chunks in your R Markdown document, making it easier to read, debug, and maintain. Each chunk should have a specific purpose, and you should avoid overly long or complex chunks.
R for Machine Learning
R, with its extensive collection of packages, has become a powerhouse for machine learning. Its flexibility, coupled with a large and active community, makes it an ideal environment for both beginners exploring basic algorithms and experts tackling complex models. This section dives into using R for machine learning, covering implementation, algorithm comparison, and project organization.
Implementing a Simple Linear Regression Model in R
Linear regression is a fundamental supervised learning algorithm used for predicting a continuous outcome variable based on one or more predictor variables. In R, we can easily implement this using the `lm()` function. Let's imagine we're predicting house prices (the outcome) based on size (the predictor). First, we'd load our data (assuming it's in a data frame called `house_data` with columns `price` and `size`).
Then, we'd fit the model: model <- lm(price ~ size, data = house_data)
summary(model)
The `summary()` function provides key statistics, including coefficients (intercept and slope), R-squared (a measure of model fit), and p-values for testing the significance of the predictors. We can then use the model to make predictions on new data: new_house_size <- data.frame(size = c(1500, 2000))
predictions <- predict(model, newdata = new_house_size)
Implementing a Simple Decision Tree Model in R
Decision trees are another popular algorithm, particularly useful for both classification and regression tasks. They create a tree-like model of decisions and their possible consequences. The `rpart` package provides a convenient way to build decision trees in R. Let's say we want to predict whether a customer will click on an ad (classification) based on features like age and income.
We'd load the data (let's call it `ad_data` with columns `click` (0 or 1), `age`, and `income`), and then: library(rpart)model <- rpart(click ~ age + income, data = ad_data, method = "class") # "class" for classification
#For regression, use method = "anova"
printcp(model) #Displays complexity parameter
plot(model, uniform=TRUE, main="Decision Tree for Ad Click Prediction")
text(model, use.n=TRUE, all=TRUE, cex=.8)
The `printcp()` function shows the complexity parameter, helping us choose the optimal tree size to avoid overfitting. The `plot()` and `text()` functions visualize the resulting tree. Predictions can then be made using `predict()`, similar to linear regression.
Comparison of Machine Learning Algorithms for Classification and Regression
Several algorithms excel at classification (predicting categorical outcomes) and regression (predicting continuous outcomes).For classification, common choices include:
- Logistic Regression: Predicts the probability of an event. Good for binary classification (two outcomes).
- Support Vector Machines (SVM): Finds the optimal hyperplane to separate data points. Effective in high-dimensional spaces.
- Random Forest: An ensemble method that combines multiple decision trees. Robust and often provides high accuracy.
- Naive Bayes: Based on Bayes' theorem, assuming feature independence. Simple and efficient.
For regression, popular options are:
- Linear Regression: Models the relationship between variables with a linear equation.
- Support Vector Regression (SVR): An extension of SVM for regression tasks.
- Random Forest Regression: Similar to random forest for classification, but for continuous outcomes.
- Decision Tree Regression: A decision tree used for predicting continuous values.
The best algorithm depends on the specific dataset and problem. Factors to consider include data size, dimensionality, linearity, and the desired level of interpretability.
Organizing a Machine Learning Project in R
A well-structured machine learning project is crucial for reproducibility and collaboration. Here's a suggested workflow:
1. Data Acquisition and Preprocessing
Gather data, clean it (handle missing values, outliers), and transform it (e.g., scaling, encoding categorical variables). Utilize packages like `dplyr` and `tidyr` for data manipulation.
2. Exploratory Data Analysis (EDA)
Visualize the data using `ggplot2` to understand patterns, relationships, and potential issues.
3. Model Selection and Training
Choose appropriate algorithms based on the problem type and data characteristics. Split the data into training and testing sets using `caret`'s `createDataPartition()` function. Train the models using relevant R packages (e.g., `glm` for logistic regression, `svm` for SVM, `randomForest` for random forests).
4. Model Evaluation
Assess model performance using metrics like accuracy, precision, recall (for classification), or RMSE, R-squared (for regression). The `caret` package provides functions for model evaluation.
5. Model Tuning and Selection
Optimize model hyperparameters using techniques like cross-validation. The `caret` package offers tools for this.
6. Deployment and Monitoring
Deploy the best-performing model and monitor its performance over time.
Debugging and Troubleshooting in R
So, you've written some awesome R code, but it's throwing errors like a toddler in a candy store. Don't worry, we've all been there. Debugging is a crucial part of the R programming journey, and mastering these techniques will save you countless hours of frustration. This section will cover common R errors, debugging tools, and strategies for writing cleaner, more error-resistant code.Debugging in R is less about avoiding errors altogether (that's nearly impossible!) and more about efficiently identifying and squashing them when they inevitably pop up.
Think of it as a detective game where you're tracking down the culprit causing your code to malfunction.
Common R Errors and Solutions, R programming language
Let's dive into some of the most frequently encountered errors in R and their fixes. Understanding these common pitfalls will make your debugging process significantly smoother.
- Object not found: This classic error appears when you try to use a variable or function that hasn't been defined or loaded. The solution? Double-check your spelling, ensure the object is in your current environment (using `ls()` to list objects), and verify that any necessary packages are loaded with `library()`. For example, if you get an error referencing `mydata`, make sure you've actually created or imported a data frame named `mydata`.
- Incorrect data types: R is picky about data types. Trying to perform an arithmetic operation on a character string, for example, will lead to an error. Use functions like `is.numeric()`, `is.character()`, and `is.factor()` to check your data types and convert them as needed using functions like `as.numeric()`, `as.character()`, etc. If you're trying to add a number to a string, you'll need to convert the string to a number first.
- Incorrect function arguments: Each function in R has specific arguments. Providing the wrong number of arguments, or arguments of the wrong type, will result in errors. Consult the function's documentation (using `?function_name`) to see the correct usage. For example, if a function expects a data frame as an argument, you can't pass it a character string.
- Missing parentheses or brackets: A simple misplaced parenthesis or bracket can throw off your entire code. Carefully review your code for any missing or mismatched parentheses or brackets.
- Index out of bounds: When accessing elements of a vector or matrix, make sure your index is within the valid range. Trying to access an element beyond the length of a vector will cause an error. Always check the dimensions of your data structures using functions like `dim()` or `length()`.
Using R's Debugging Tools
R provides several built-in tools to help you track down errors. These tools allow you to step through your code line by line, inspect variable values, and identify the exact source of the problem.The most common debugging tool is the `debug()` function. You place `debug()` before a function call, and then R enters interactive debugging mode when the function is called.
You can then step through the code using commands like `n` (next), `s` (step into), `c` (continue), and `Q` (quit). This allows you to see the state of your variables at each step and pinpoint where the error occurs. The `browser()` function works similarly and can be placed anywhere in your code to start debugging.Another helpful tool is the `traceback()` function.
When an error occurs, `traceback()` displays the sequence of function calls that led to the error, helping you trace the error back to its source.
Strategies for Writing Clean and Well-Documented Code
Proactive measures are key to minimizing debugging headaches. Writing clean, well-documented code makes it much easier to spot errors and understand your code's logic, both now and in the future.
- Use meaningful variable names: Avoid cryptic abbreviations; use descriptive names that clearly indicate the variable's purpose. For example, `customer_age` is better than `ca`.
- Add comments: Explain complex logic or non-obvious code sections with comments. This makes your code easier to understand and maintain.
- Break down complex tasks into smaller functions: This improves code readability and makes it easier to identify and fix errors in individual functions.
- Use consistent indentation: Consistent indentation improves code readability and helps you spot structural errors.
- Test your code incrementally: Don't write a huge chunk of code and then try to debug it all at once. Test small sections as you go to catch errors early.
Last Point
So, there you have it – a whirlwind tour through the exciting world of R! From its origins to its current capabilities, we’ve covered a lot of ground. Remember, mastering R is a journey, not a sprint. Keep practicing, exploring new packages, and most importantly, have fun analyzing data! The power to unlock insights from information is in your hands now.
Go forth and analyze!
Quick FAQs
What are some good resources for learning R beyond this guide?
Check out websites like DataCamp, Coursera, and edX for online courses. There are also tons of helpful tutorials and documentation on the official R website and through community forums.
Is R difficult to learn?
Like any programming language, R has a learning curve. But with consistent practice and access to good resources, it's definitely manageable. Start with the basics and gradually build your skills.
What's the difference between R and Python for data science?
Both are popular choices! R excels in statistical computing and data visualization, while Python offers broader general-purpose programming capabilities. Many data scientists use both!
How much math do I need to know to use R effectively?
A basic understanding of statistics is helpful, especially for statistical modeling. However, you can still use R for data manipulation and visualization without being a math whiz.
Where can I find R packages?
The Comprehensive R Archive Network (CRAN) is the primary repository for R packages. You can install them directly from within R using the `install.packages()` function.

